cloud

VHP4Safety Cloud catalog
Below you find an overview of services available in the context of the VHP4Safety Platform. Additional services have been suggested and users can request additional services.
AOP-Builder
A tool that supports Adverse Outcome Pathway (AOP) development by extracting key scientific entities and their relationships from literature. [more information]
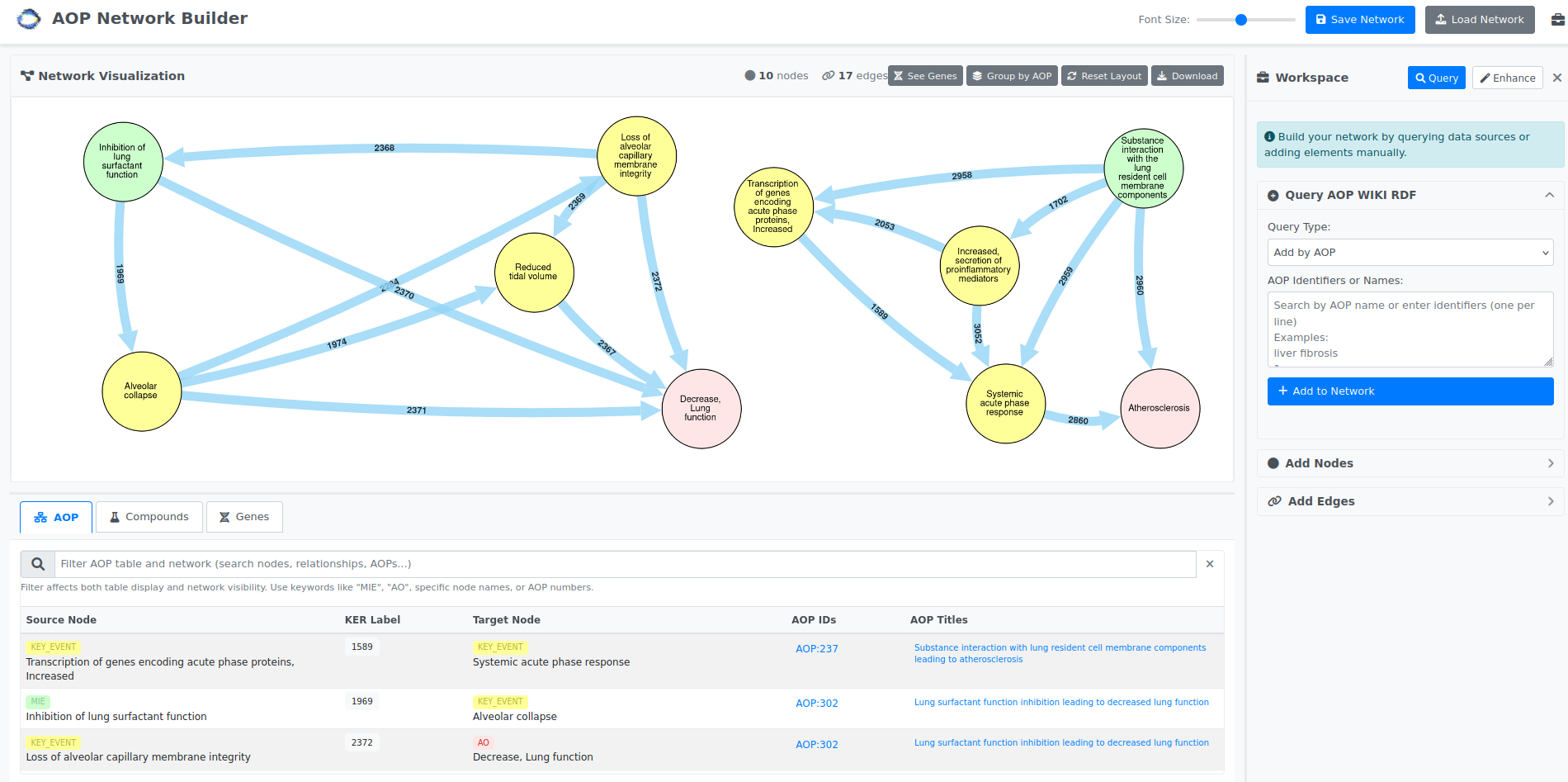
AOP-Suite
A web-based tool for building, visualizing and analyzing Adverse Outcome Pathway (AOP) networks. [more information]
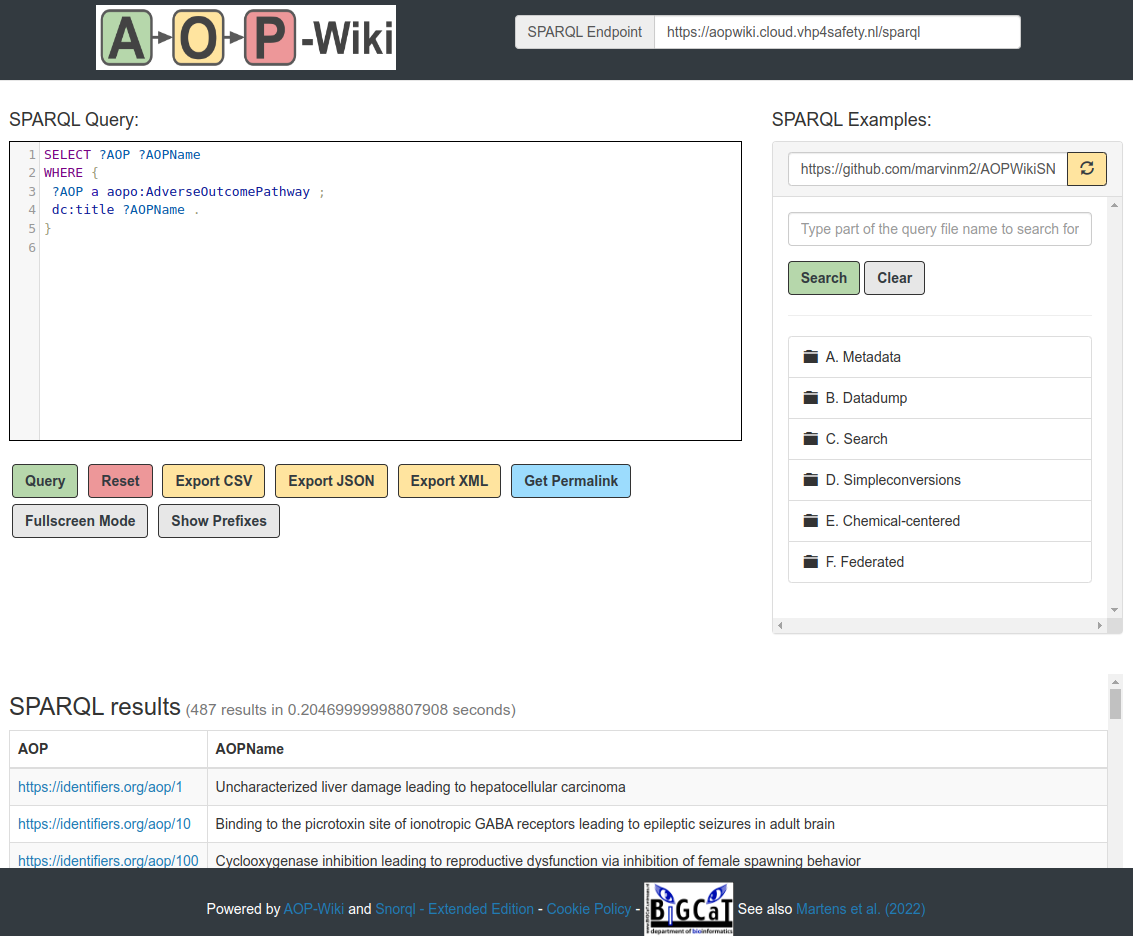
AOP-Wiki Snorql UI
A graphical interface for creating, preloading and exporting SPARQL queries tailored to your usecases. [more information]
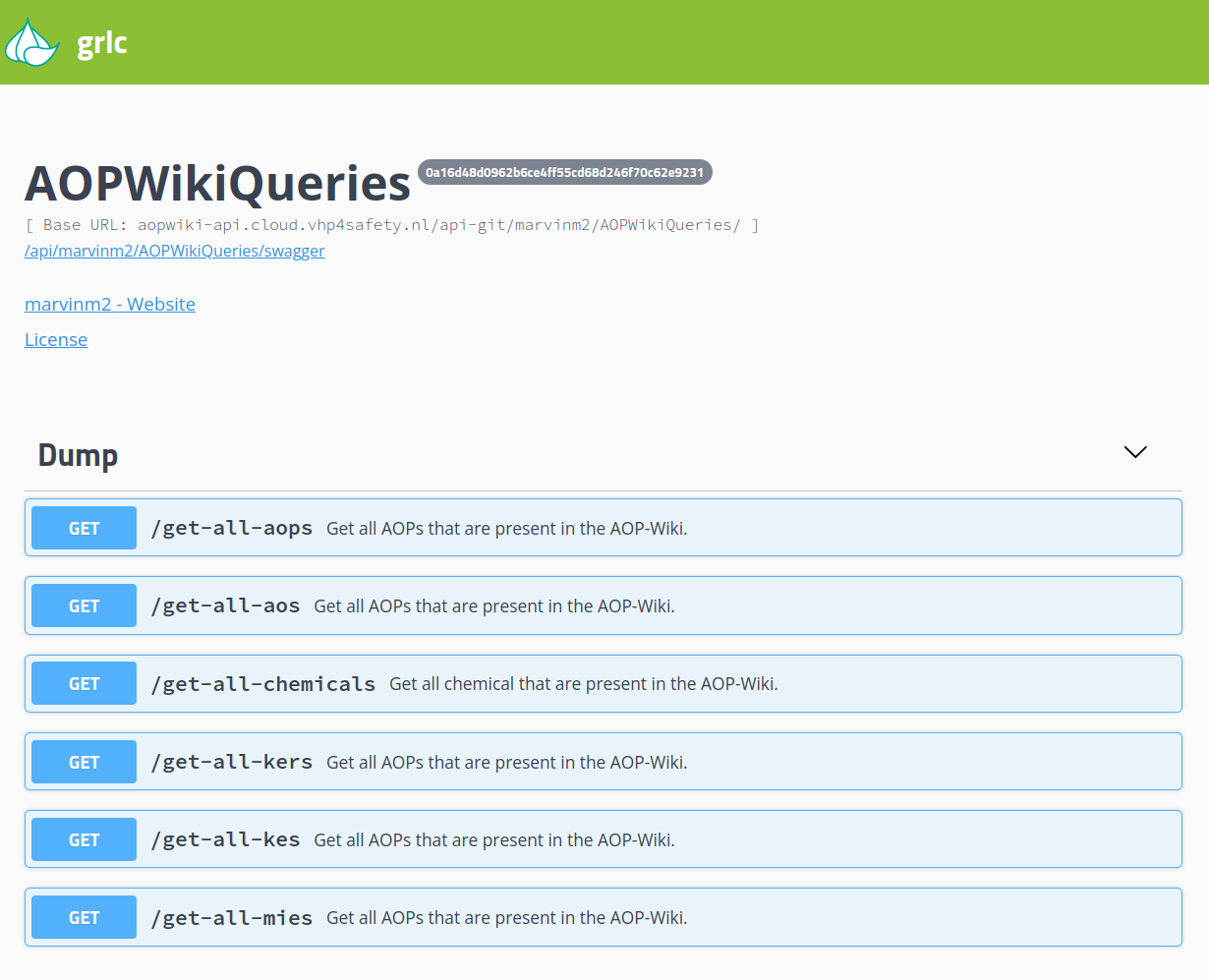
AOP-Wiki API
A REST API for creating and accessing content in the AOP-Wiki database. [more information]

ArrayAnalysis
A user-interface for preprocessing and statistically analyzing RNA-sequencing and microarray transcriptomics data. [more information]
ASReview
Open-source active-learning software for transparent and efficient systematic literature reviews. [more information]

BioModels
A repository dedicated to the inventory and acquisition of high-quality mathematical models pertaining to biological and biomedical systems. [more information]
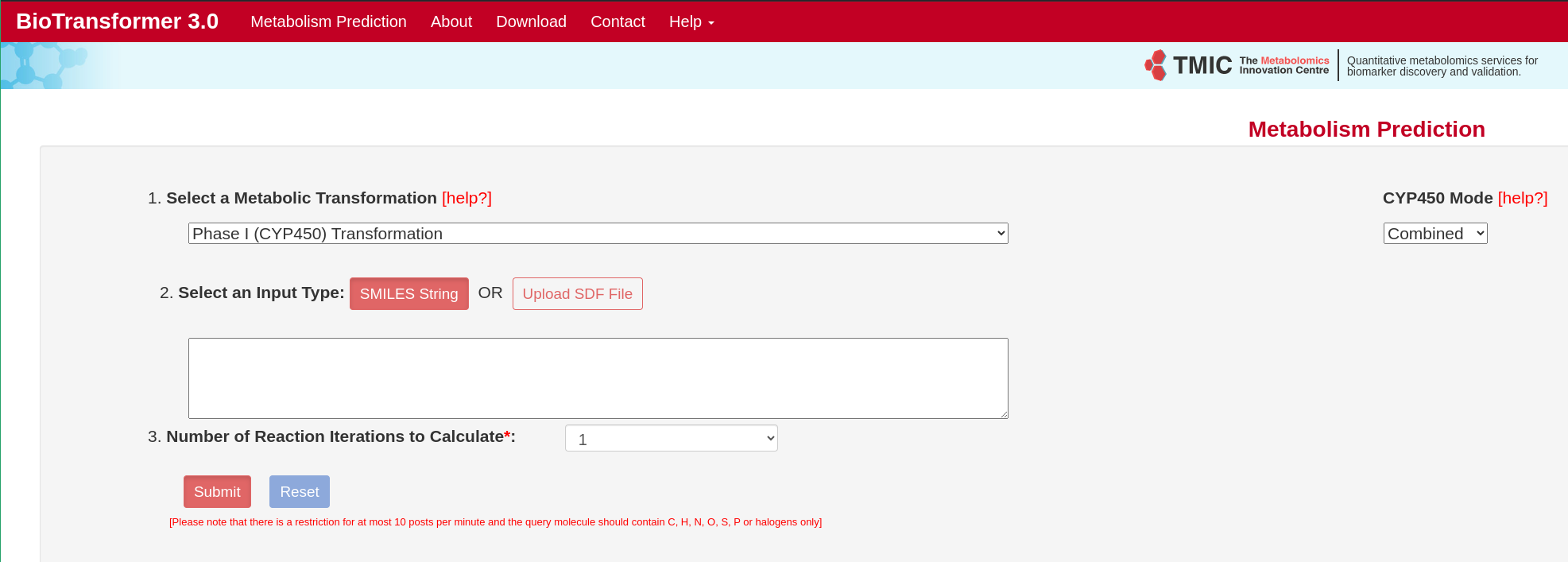
BioTransformer
A software tool for predicting metabolites from SMILES-formatted chemical inputs. [more information]
BMDExpress-3
A software tool for high-dimensional dose-response modeling and analysis. [more information]

BridgeDb
A framework for mapping identifiers across biological databases and related sources. [more information]
The Chemistry Development Kit
A Java library for chem- and bioinformatics, with functionality to read/write chemical structures, calculate molecular descriptors, physicochemical properties, substructure and similarity searching, and much more. [more information]
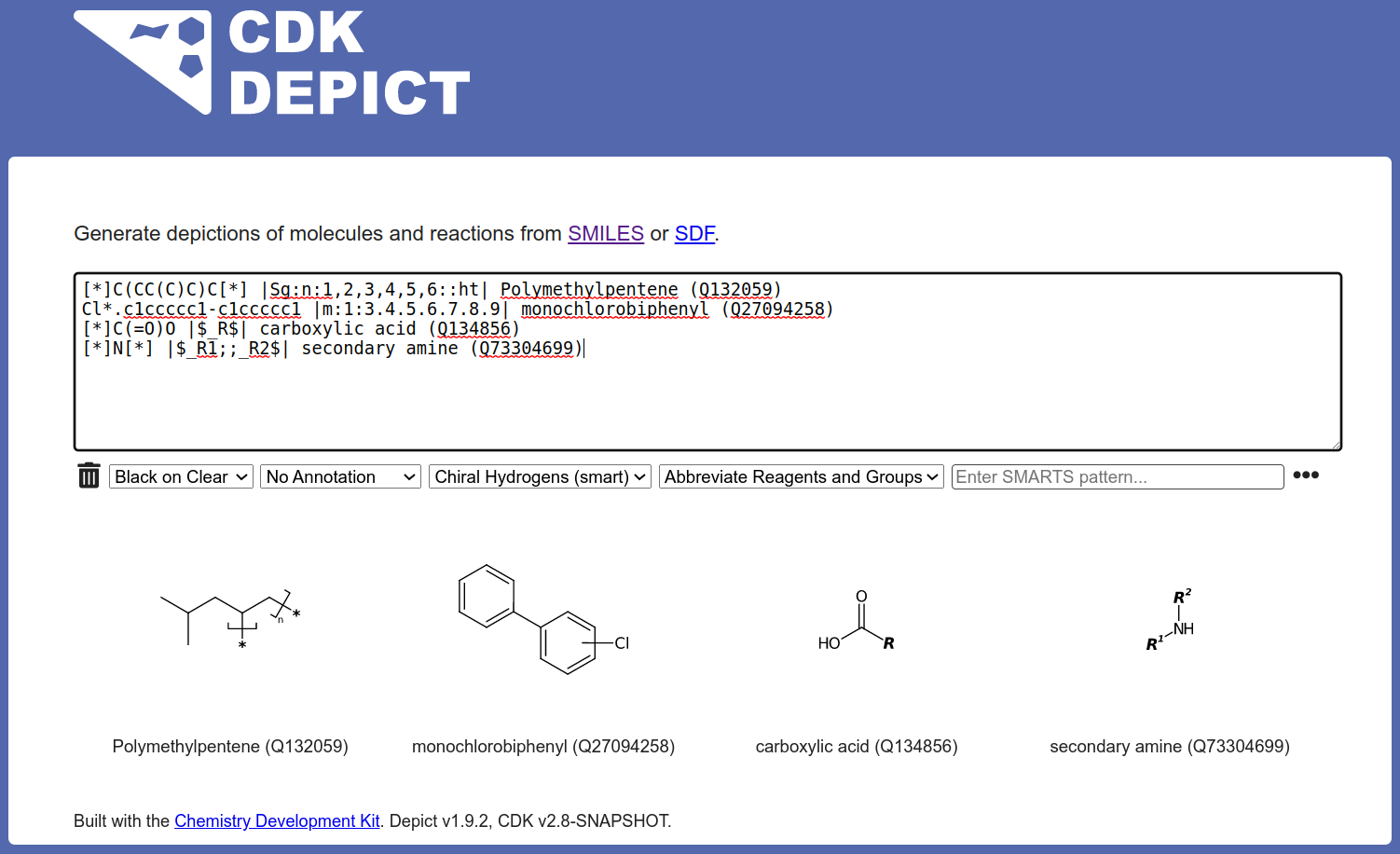
CDK Depict
A webservice for generating chemical structure images from SMILES inputs. [more information]

CellDesigner
An intuitive user interface for creating gene-regulatory and biochemical network diagrams. [more information]

CompTox
An online database for searching chemical, toxicity and exposure information. [more information]

COPASI
An open-source software for simulating, analyzing and visualizing biochemical networks using a wide range of methods. [more information]
cpLogD
An online model for predicting water-octanol distribution coefficient (logD) of chemical compounds with confidence estimates. [more information]
Danish (Q)SAR Database
A freely available (Q)SAR tool for substance evaluations that includes estimates from more than 200 (Q)SARs covering a wide range of hazardous properties relevant for human health and the environment. [more information]

DECIMER
An online service for automatically extracting chemical structure depictiions from PDFs and images. [more information]
NIH Dietary Supplement Label Database
An online database from the NIH Office of Dietary Supplements that lets you explore information from more than 200,000 dietary supplement labels sold in the United States. [more information]

European Medicines Agency Documents
An online repository for inventarisation of official reports from the European Medicines Agency concerning chemical evaluation and safety monitoring. [more information]
Endocrine Disruptome
An open source, user friendly and web based prediction tool that uses molecular docking to predict binding of compounds to 14 different nuclear receptors. [more information]

FAIRDOMHub
A web-accessible registry for managing, sharing and organising research data and research assets. [more information]

Fairspace
An open source RDF-based data management platform for importing and exporting research-relevant metadata [more information]

Farmacotherapeutisch Kompas
A Dutch online resource for gathering data on the pharmacology, adverse effects, and prices of all pharmaceutical products available in the Netherlands. [more information]
Flame
An open source web application for building predictive models from biologically annotated chemical structures using predefined workflows. [more information]
Google Search
Online search engine for Google. [more information]
Google Scholar
An online search engine for (scientific) literature. [more information]

Joint Research Centre Data Catalogue
An in-house data inventory produced by the Joint Research Centre (JRC) for provision of independent scientific advice and support to policies of the European Union. [more information]
Llemy
An online tool for answering questions about lipid metabolism and related toxicology/biology using MINERVA pathway knowledge, live web research and AI-generated summaries. [more information]
MCT8 Docking for MIE Discovery
A Python-based notebook for identifying potential MCT8 inhibitors and related molecular events linked to developmental thyroid toxicity from maternal thyroid hormone deprivation. [more information]
MolAOP analyser
An online service for analyzing transcriptomic data in the context of molecular Adverse Outcome Pathways (AOPs), supporting data upload or demo datasets for gene selection, visualization and Key Event (KE) enrichment analysis. [more information]
Molecular AOP Builder
A web application for mapping Key Events from Adverse Outcome Pathways to WikiPathways and Gene Ontology Biological Process terms, using intelligent suggestions based on gene overlap, text similarity, and BioBERT semantic embeddings. [more information]

The OECD QSAR Toolbox
A software for supporting chemical hazard assessment and enhancing mechanistic knowledge of substances in a cost-effective way. [more information]
ONTOX Physiological Maps
Maps for illustrating biological processes behind toxicological risks using a standardized graphical notation. [more information]
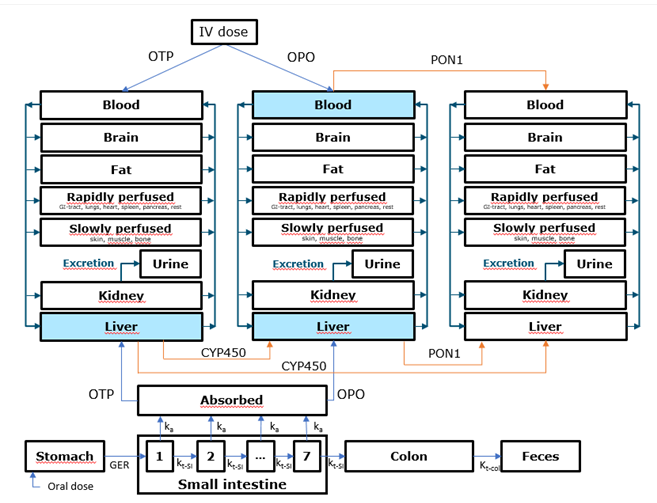
OP PBK Model
An online physiologically based pharmacokinetic model for organophosphates [more information]
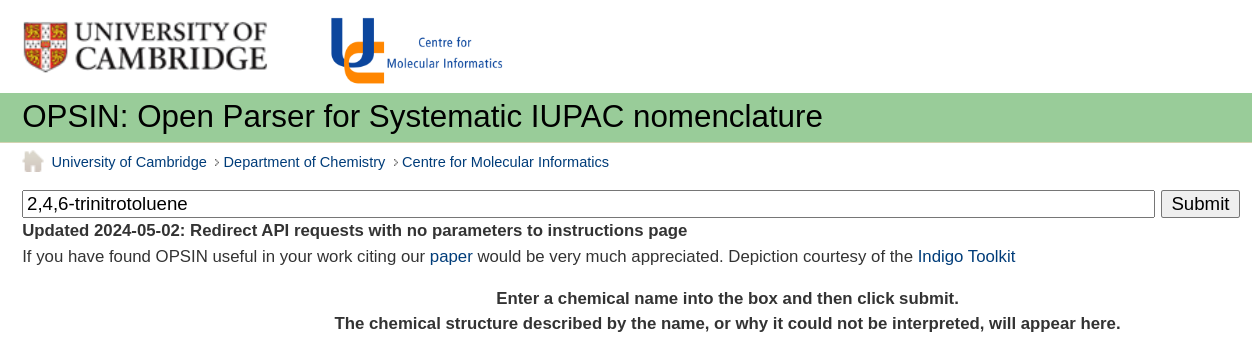
OPSIN: Open Parser for Systematic IUPAC nomenclature
An online service for parsing chemical IUPAC names into a machine-readable chemical identifiers. [more information]

O-QT — OECD QSAR Toolbox AI Assistant
An online assistant for using the OECD QSAR Toolbox to analyze chemicals, assess hazards and generate AI-based summary reports. [more information]
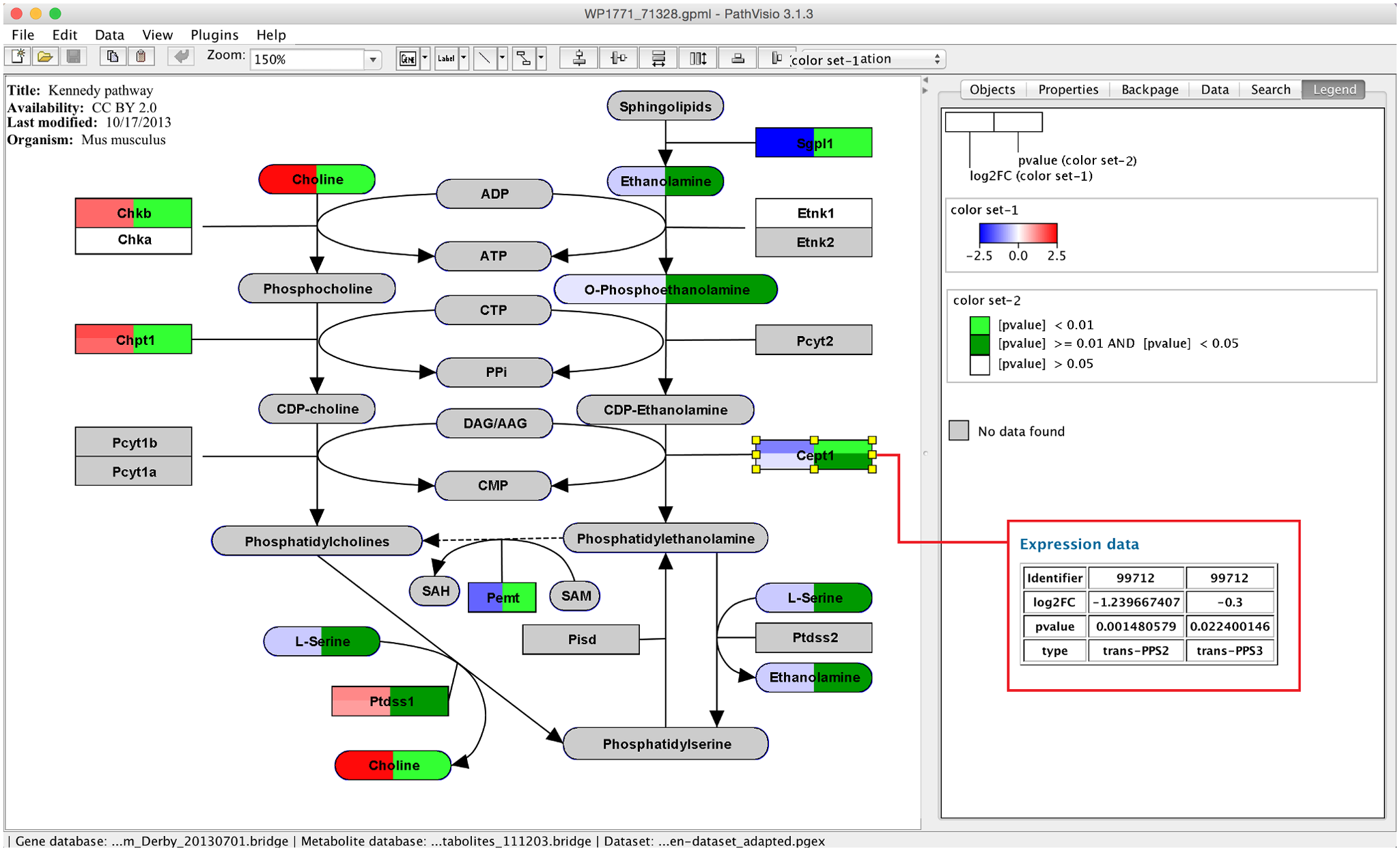
PathVisio
Desktop software to visualize, edit, and draw new pathways, which can be exported in the GPML format, for example, to WikiPathways. [more information]
qAOP-App
An interactive tool for visualizing and predicting key events (KEs) and adverse outcomes (AOs) over time using ODE-based qAOP models, illustrated with VHP4Safety case studies. [more information]

QSPRpred
An online tool for chemical activity prediction for various endpoints using target-specific and pretrained QSAR models. [more information]

R-ODAF Shiny
An R Shiny interface for the R-ODAF framework that provides standardised RNA-seq differential expression analysis for regulatory toxicology applications. [more information]
SOM Prediction
A webservice for prediction of protein-structure and reactivity based (P450) site-of-metabolism. [more information]
SysRev
An online (proprietary) service for literature review, data extraction and systematic review; consider ASReview as the preferred open-source alternative. [more information]

ToxTempAssistant
An online tool for harmonizing in vitro toxicity test methods using Large Language Models, based on the ToxTemp template where it extracts details from protocols, publications or lab notes to create structured draft responses. [more information]
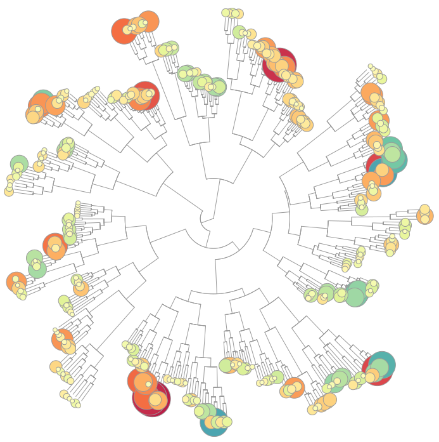
TXG-MAPr
An online application for implementation of weighted gene co-expression network analysis (WGCNA) derived from publicly available transcriptomic datasets from the TG-GATEs database. [more information]
VHP4Safety Glossary
A collection of ontology with names, abrreviations and definitions of the terminology that is used within the VHP4Safety project. [more information]
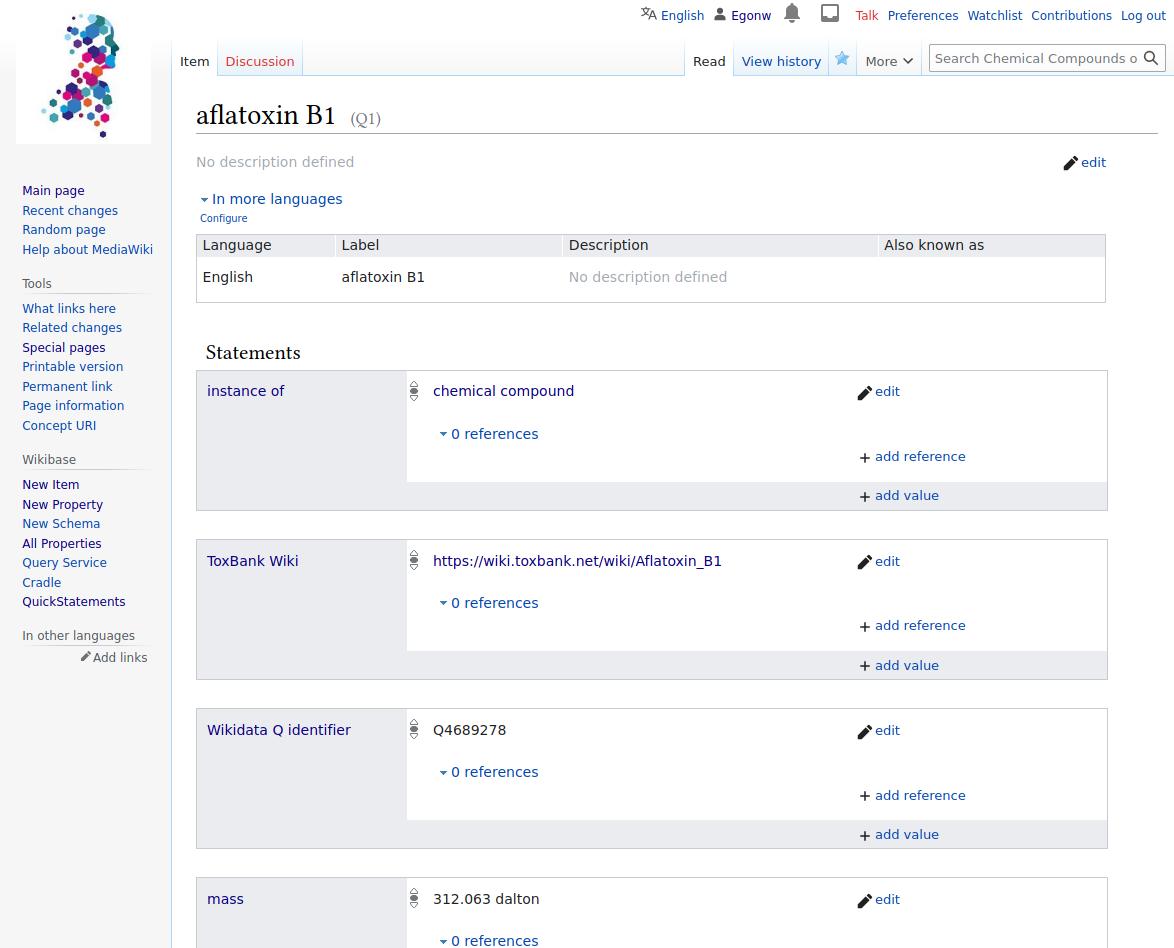
VHP4Safety Wikibase
A Wikibase instance for storing and presenting information about toxic, safe and potentially toxic compounds related to the VHP4Safety project. [more information]
WikiPathways - AOP Portal
An Adverse Outcome Pathway (AOP) portal on WikiPathways for exploring the molecular basis of AOPs and their key events (KEs). [more information]
xploreaop
A web application for visualization of Adverse Outcome Pathway (AOP) networks and interactive exploration of AOPs for two liver outcomes. [more information]
Funding
VHP4Safety – the Virtual Human Platform for safety assessment project NWA 1292.19.272 is part of the NWA research program ‘Research along Routes by Consortia (ORC)’, which is funded by the Netherlands Organization for Scientific Research (NWO). The project started on June 1, 2021 with a budget of over 10 million Euros and will last for the duration of 5 years.